Gene Regulatory Interaction Network Simulator (GRiNS)
A Python library for simulating gene regulatory networks (GRNs) using parameter-agnostic frameworks like RACIPE and Ising formalism, with GPU acceleration and efficient ODE solving.
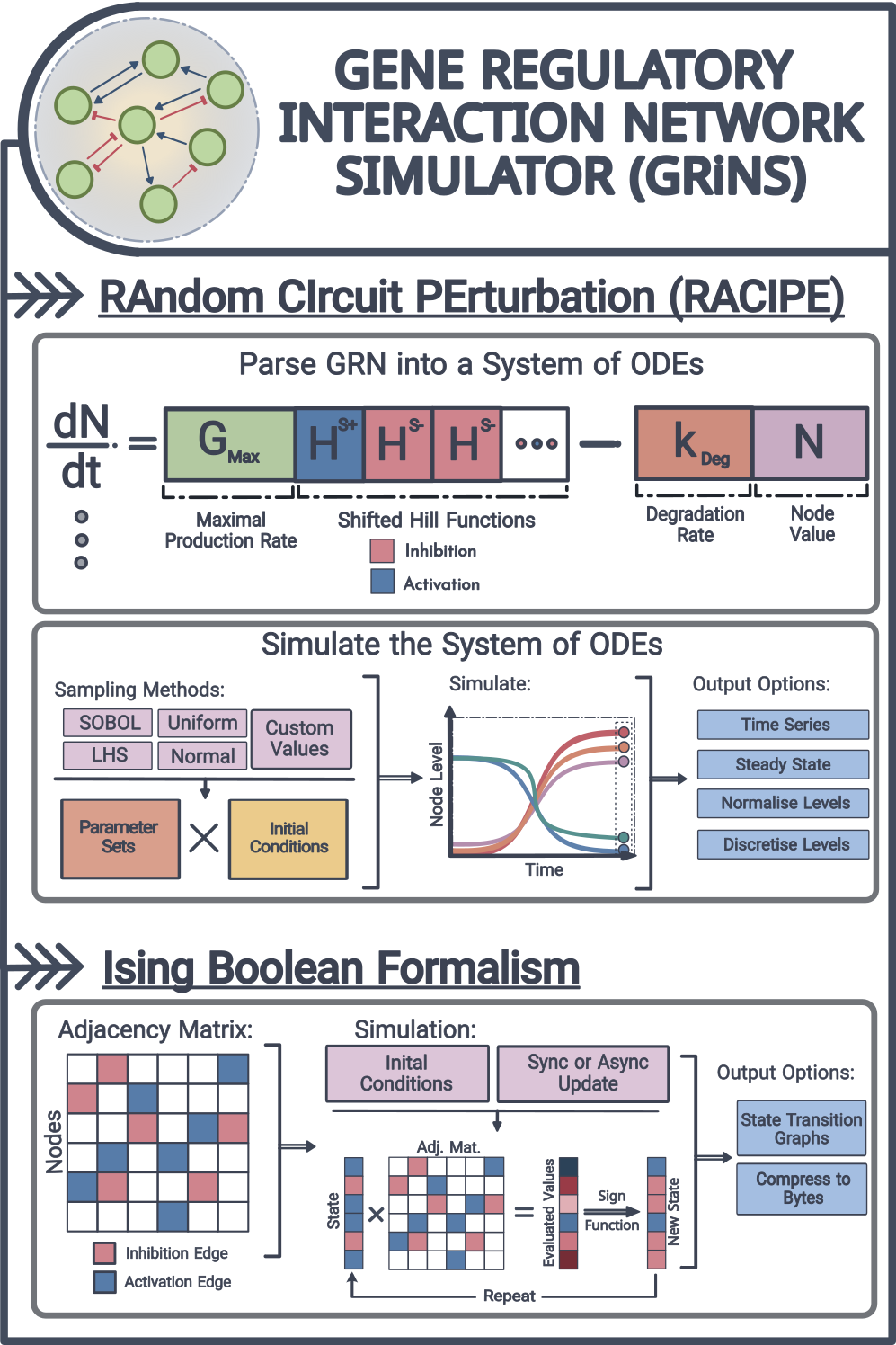
Modeling gene regulatory networks (GRNs) is essential for understanding cellular processes, but parameterizing these networks becomes increasingly difficult as they scale. This Python library provides a simulation framework that unifies parameter-agnostic approaches, including RACIPE and Ising formalism, into a single, flexible tool.
Key Features
- Simulation Frameworks: Supports both ODE-based (RACIPE) and coarse-grained (Ising formalism) methods for studying GRN dynamics.
- Parameter-Agnostic Modeling: Translates network topology into mathematical models without requiring detailed parameter tuning.
- Scalable Computation: Uses the Jax ecosystem for GPU acceleration and Diffrax for efficient ODE solving.
- Data Processing Tools: Provides normalization and discretization functions to standardize simulation outputs for downstream analysis.
Installation
GPU Version Installation (Recommended)
For optimal performance, it is recommended to install the GPU-accelerated version of the library. This version leverages CUDA for faster computations, making it well-suited for large-scale simulations. If you have a compatible NVIDIA GPU (refer to Jax Installation), install the library with:
CPU Version Installation
If you do not have a compatible GPU, you can install the CPU version instead:
Compared to the GPU version, the CPU version will be slower, especially for large simulations.
Citation
Please cite this package if you have used it.
@misc{harlapur2025grinspythonlibrarysimulating,
title={GRiNS: A Python Library for Simulating Gene Regulatory Network Dynamics},
author={Pradyumna Harlapur and Harshavardhan B V and Mohit Kumar Jolly},
year={2025},
eprint={2503.18356},
archivePrefix={arXiv},
primaryClass={q-bio.QM},
url={https://arxiv.org/abs/2503.18356},
}